| Version 83 (modified by , 8 years ago) (diff) |
|---|
How to add a sample in NUISANCE
This is an informal step-by-step guide on how to add samples into the NUISANCE framework. Most of this can be derived by looking at the recently implemented measurements for up-to-date references.
For this tutorial I'll be adding the T2K CC1π+ H2O data. The data comes from the T2K site and arxiv. The data is supplied both in a ROOT file and a number of .csv files: I'll be using the ROOT file here but very similar steps apply when using .csv or .txt files.
After implementation we'll be able to make data/MC comparisons to the T2K CC1π+ H2O data for NEUT, GENIE, GiBUU and NuWro, all in the same framework. We'll get MC predictions, χ2 values, interaction mode contributions, and more using a consistent signal definition, guaranteed the same across the generators.
NUISANCE is currently fully written in C++03 and we would appreciate future contributions to adhere to this standard.
If you've been using this guide we'd much appreciate feedback, thank you.
Author: Clarence Wret
Date: January 2017
Versions: NUISANCE v1r0, NEUT 5.3.6, arxiv 1605.07964v2.pdf
Wiki content
- How to add a sample in NUISANCE
- Examining the data and choosing distributions
- Preparing structure for a sample
- Coding up a sample
- Add the sample to the CMake process
- Make NUISANCE aware of the sample
- Recompiling NUISANCE with the new sample
- Running NUISANCE with the new sample
- The final implementation
- What about 2D distributions?
Examining the data and choosing distributions
Finding the neutrino flux and generating events
The first issue at hand is to find the flux for the experiment. If we don't have this, we cant make a generator prediction--unless the measurement is a total cross-section without any phase space cuts (in which case you should probably cast a suspicious eye).
A quick search through the arxiv document points us to reference [12]. It is also in our flux list.
I then generate events in NEUT 5.3.6 with a suitable card-file, see our NEUT guide on how to do this. The procedure is very similar for other generators too. We need the correct target (water) and flux, and perform the model selections we want (e.g. Rein-Sehgal or Berger-Sehgal coherent model).
Aim to generate around 1M events with all interaction modes turned on. This way we make sure to get all interaction modes into the topologically defined cross-section. We get a small amount of CCQE events which excite a pion from the outgoing nucleon interacting in the nucleus to kick out a pion, leading to a CCQE+1π+ ~ CC1π+ final state, which is classified signal for this particular sample.
Choosing a cross-section distribution
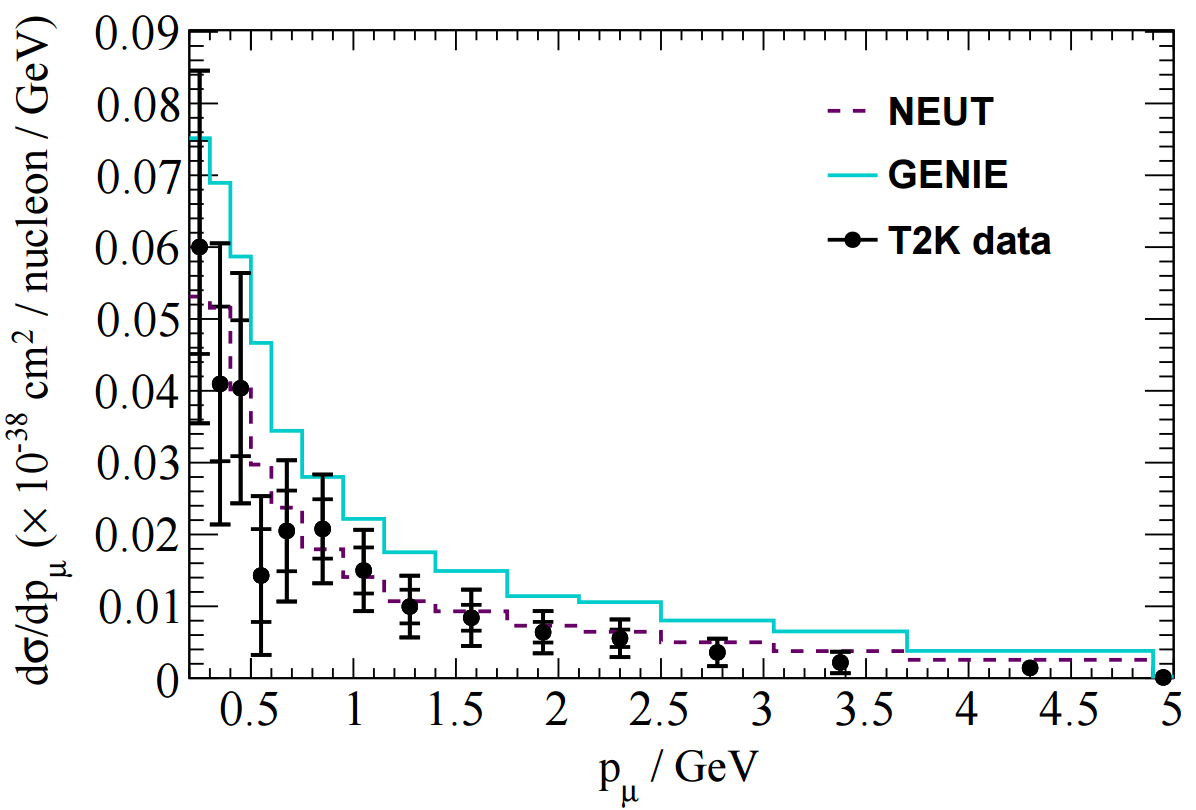
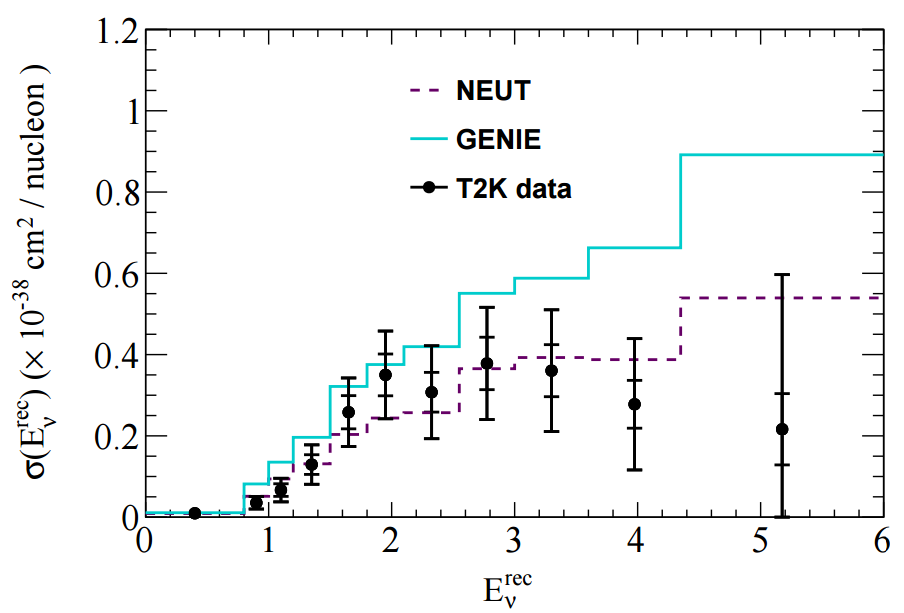
The T2K CC1π+ H2O data release contains various distributions in FIG 4. In this tutorial I'll look at adding one kinematic distribution and one derived distribution: pµ and Erecν, shown below.
In NUISANCE we try to add all available distributions from a publication. However, some distributions will have detector effects "folded" into them (i.e. they will be raw detector-level data). We can only use these if there is some method which bring detector-level variables (e.g. pµ seen in the detector) to truth-level variable (e.g. pµ seen after correcting for the detector effects).
Preparing structure for a sample
NUISANCE is designed to easily allow adding new samples.
Each experimental measurement is its own class. To simplify including new samples we supply a base class for 1D (Measurement1D) and 2D (Measurement2D) measurements, which in turn inherits from the virtual base class (MeasurementBase).
These MeasurementBase classes are looped over from the executables and data/MC comparisons are the result, calling functions like MeasurementBase::Reconfigure, MeasurementBase::FillEventVariables, MeasurementBase::isSignal and MeasurementBase::GetLikelihood.
A class has to at least inherit from MeasurementBase and implement the necessary methods to be "looped over". The recommended method is to use and expand the supplied Measurement1D and Measurement2D classes; anything else may require expert knowledge and won't be covered here.
The inheritance tree is simple and goes Specific_Measurement -> MeasurementXD -> MeasurementBase
Naming the sample
Some automatic processing is done on loading up the samples to set up generator scaling factors, χ2 calculations and so on. These are simple string comparisons done in the base class constructors, but do place responsibility on the user.
The structure is Experiment_Measurement_Target_DataType_DimensionVar_Neutrino:
Experimentis the experiment (for us T2K)Measurementis a suitable name for the cross-section (for us CC1pip)Targetis the interaction target (for us H2O)DataTypeis the measurement type, eitherEvtfor an event rate measurement orXSecfor a cross-section measurement (for usXSec)DimensionVaris 1D or 2D followed by a suitable name for the dependent variable (for us1Dpmu)Neutrinois the neutrino type (for usnu)
Out of these, string comparisons are only made on DataType and DimensionVar. The other identifiers exist to adhere to some standard and keep sample naming tidy and consistent.
What to do:
Following the above convention, we end up with T2K_CC1pip_H2O_XSec_1Dpmu_nu and T2K_CC1pip_H2O_XSec_1DEnuMB_nu.
This is a sufficient name: there is no ambiguity what the class describes and there is no way of confusing it with other classes in NUISANCE.
Placing the sample in a directory
As with many packages, NUISANCE has source code in the src directory. We have a directory for each experiment inside src, e.g. src/T2K, src/MINERvA, and so on.
The applications using the src files go in the app directory. However, including a new sample does not involve changing anything there, so let's ignore it for the purpose of this tutorial.
The new files themselves for the new samples should be identical (or very similar) to the sample name to avoid confusion.
What to do:
Make the new files src/T2K/T2K_CC1pip_H2O_XSec_1Dpmu_nu.cxx and src/T2K/T2K_CC1pip_H2O_1Dpmu_nu.h. These are the implementation and header files for the new measurement and similar goes for the 1DEnuMB implementation.
Placing the data in a directory
The data for the measurement goes into the data directory.
As with the src directory we have sub-directories for each experiment, e.g. data/T2K and data/MINERvA.
Furthermore, we specify another directory for the measurement topology to avoid confusion, e.g. data/T2K/CC1pip for our sample.
In some cases we might have the same topology defining the cross-section but for a different interaction target. In this case we add another sub-directory for the target, e.g. data/T2K/CC1pip/H2O for us.
What to do:
Put the ROOT file from the data release into the data/T2K/CC1pip/H2O directory.
Coding up a sample
Now that we have the rough structure set-up, we can start writing some code.
Writing the header
The T2K CC1π+ H2O data are all 1D distributions. The new classes should therefore inherit from the Measurement1D base class, as mentioned earlier. The Measurement1D class is implemented in src/FitBase/Measurement1D.cxx.
NUISANCE requires a constructor and destructor for the class, and we'll need to overload MeasurementBase methods which define the dependent variable(s) (pµ and Erecν in our case) and what our signal is (CC interaction with one muon and one positive pion with no other pions or mesons and any number of nucleons in our case). The MeasurementBase functions which we need to overload are MeasurementBase::isSignal(FitEvent *event) and MeasurementBase::FillEventVariables(FitEvent *event).
The FitEvent class is an object which contains information about one single event: all the particles, their kinematics and their status before and after the interaction, the interaction channel(s) which produced the final state particles, possible secondary interactions, the interaction target, and so on. The FitEvent class is implemented in src/FitBase/FitEvent.cxx and is the generator-independent "common event format" which NUISANCE uses for the generator events.
We also note that the src/T2K directory contains T2K_SignalDef.cxx. This extends the SignalDef namespace which holds many of the signal definitions for the classes. SignalDef itself lives in src/Utils/SignalDef.cxx, but may see additions from experiment folders such as src/T2K/T2K_SignalDef.cxx.
If you're implementing multiple distributions for the same measurement (e.g. dσ/dpµ and dσ/dcosθµ) we recommend adding the signal definition to the namespace rather than directly in the isSignal(FitEvent *event) function in the class implementation. We'll talk about implementing a signal definition later.
What to do: Our header file now looks like:
#ifndef T2K_CC1PIP_H2O_XSEC_1DPMU_NU_H_SEEN
#define T2K_CC1PIP_H2O_XSEC_1DPMU_NU_H_SEEN
#include "Measurement1D.h"
#include "T2K_SignalDef.h"
class T2K_CC1pip_H2O_XSec_1Dpmu_nu : public Measurement1D {
public:
T2K_CC1pip_H2O_XSec_1Dpmu_nu(std::string inputfile,
FitWeight *rw, std::string type, std::string fakeDataFile);
virtual ~T2K_CC1pip_H2O_XSec_1Dpmu_nu() {};
void FillEventVariables(FitEvent *event);
bool isSignal(FitEvent *event);
private:
};
#endif
where we've copied the constructor arguments from some other class we're using as a template. I'll elaborate more on this below.
Note the simple structure for the sample: we really only need to set up three functions. The inheritance from Measurement1D and helper function within it does most of the heavy lifting.
Writing the constructor
NUISANCE loads samples through FCN/SampleList.cxx in the function SampleUtils::LoadSample. This function makes a std::list of MeasurementBase pointers which it later loops over in calls from the executables.
Most of the automated setting up of histograms, titles, and so on is done in Measurement1D::SetupMeasurement(std::string inputfile, std::string type, FitWeight *rw, std::string fakeDataFile). It hence makes sense to have a class constructor which passes all of these on to SetupMeasurement.
The arguments to Measurement1D::SetupMeasurement are:
inputfile: the location of the input file containing generated eventstype: the type of comparison we want to make with this samplerw: the reweighting engine which may or may not be used to reweight the samplefakeDataFile: the location of a fake-data input file which is to be used as data instead of the actual data for fake-data studies
which currently all have to be passed. In reality, both rw and fakeDataFile do nothing in SetupMeasurement but are left for legacy reasons. We will likely change this soon!
Optional:
Specifying type enables configuration of the class on run-time in the constructor. type is a std::string that gets read from the card file specified by the user at the end of each line.
This is useful if the same measurement is made but with different kinematic cuts, e.g. ANL CC1π+1p has W < 1.4, 1.6 and no cut data. We could pass arguments in our card file to load different data and cut at different W values on run-time without having to add in a new class for each W cut.
To get the type running smoothly you need to changed the Measurement1D protected members fDefaultTypes and fAllowedTypes to whatever strings you want to accept in the card file, whilst also including the defaults ("FIX, FREE, SHAPE"). type gets checked against the fAllowedTypes in the SetupMeasurement to ensure a consistent and expected loading.
Example: I want to make ANL CC1π+1p accept W14 as an argument so I can cut on this variable and check how the generator performs over different W cuts vs data.
In the constructor I'll need to add:
fDefaultTypes = "FIX/DIAG";
fAllowedTypes = "FIX,FREE,SHAPE/DIAG/W14";
if (type.find("W14") != std::string::npos) {
...
Do different stuff when I want W14 data + cuts
e.g. set a bool or double to remember what settings
I'm running with, load up different data,
different covariance matrix, and so on
...
} else {
...
Do some other stuff when I don't want W14 data + cuts
...
}
I then act accordingly in isSignal and FillEventVariables (e.g. fill histogram only when W < 1.4, only count signal when W < 1.4). More on these two functions come further further donw.
When I want to run with this new setting I first recompile and then edit my card file to
sample ANL_CC1ppip_XSec_1DEnu_nu NEUT:MY_NEUT_FILE_LOCATION.root W14
which should' load up whatever I specified when writing W14 in the implementation file.
Looking at the base classes which we are inheriting our sample from (Measurement1D and MeasurementBase, both in src/FitBase), we minimally need to set:
std::string fName: The name of the samplestd::string fPlotTitles: The x and y axes of the sampledouble EnuMin: The minimum neutrino energy in the sample signal definitiondouble EnuMax: The maximum neutrino energy in the sample signal definitionTH1D *fDataHist: The data histogram scanned from the provided inputTH1D *fMCHist: The generator histogram binning with same units asfDataHistTH1D *fMCStat: The generator histogram without any scaling (i.e. Nevents)TH1D *fMCFine: The generator histogram with same units asfDataHistbut finer binningTH1 **fMCHist_PDF: The mode-by-mode generator histogramsTMatrixDSym *fFullCovar: The covariance matrixTMatrixDSym *covar: The inverted covariance matrix (bad naming, sorry!)double fScaleFactor: The scaling factor we need to go from a event rate to a cross-section (Nevt(pµ) -> dSig(pµ))
We can set most of these by calling helper functions in Measurement1D, such as
Measurement1D::SetupMeasurement: checks the naming of the sample and sets up the protected/private members, sets up the input generator processing for the sample, sets the fit optionsMeasurement1D::SetDataValues: sets the data histogram from a given fileMeasurement1D::SetCovarMatrix: sets up the covariance matrices needed to calculate likelihoodsMeasurement1D::SetupDefaultHist: sets all the generator histograms and their auxillary histograms (e.g. mode-by-mode generator histograms)
or similar functions, e.g. calling Measurement1D::SetDataFromFile instead of Measurement1D::SetDataValues. Feel free to add reasonable setter functions to the base classes if you can't find a suitable one.
Please note that the order in which the helper functions are called is crucial to proper operation. E.g. Measurement1D::SetupDefaultHist relies on fDataHist having been set and a crash is likely if it is called before then.
Current NUISANCE (v1r0) behaviour requires fName, fPlotTitles, EnuMin, EnuMax and fScaleFactor to be set by user in the implementation file. These are the only variables that need to be supplied by the user in the constructor, granted above helper functions are used.
For non-expert use, it's sufficient to just copy the fScaleFactor variable as (fEventHist->Integral("width")*1E-38)/double(fNEvents)/TotalIntegratedFlux("width");. This number is applied to the event rate histogram to take us to a cross-section histogram in Measurement1D::ScaleEvents.
If the cross-section is not in cm2/nucleon but instead cm2/CH (or any other molecule) you will have to scale the fScaleFactor by the number of available target nucleons.
For CCQE, νµ interactions only occur off neutrons, which we have 6 of in CH. But the total number of nucleons in CH is 13. So if an experiment reports their cross-section in cm2/CH for CCQE νµ and our generator output is cm2/nucleon we need to do fScaleFactor*=13./6..
If instead an experiment measures a CC1π+ cross-section in cm^2^/CH, we scale by a factor 13 because CC1π+ happens on both neutrons and protons.
It's possible that these will be automated in the future, so please look at recently implemented samples for up-to-date references.
This is where we are sensitive to the the input data format. If using a .csv or .txt file instead, Measurement1D::SetDataValues might be a better option.
Please look through the Measurement1D::SetData* functions and decide on which works best for you, or extend the current implementations to match your requirements and/or contact us
What to do:
The data and covariance matrix are available in a ROOT file for the sample, so I used Measurement1D::SetDataFromFile to set up the data and Measurement1D::SetCovarFromDataFile to set up the covariances.
fName and the other std::strings were set to reasonable names and fScaleFactor was set to the default mentioned above.
The constructor for the class now looks like:
#include "T2K_CC1pip_H2O_XSec_1Dpmu_nu.h"
// The muon momentum
//********************************************************************
T2K_CC1pip_H2O_XSec_1Dpmu_nu::T2K_CC1pip_H2O_XSec_1Dpmu_nu(
std::string inputfile, FitWeight *rw,
std::string type, std::string fakeDataFile){
//********************************************************************
fName = "T2K_CC1pip_H2O_XSec_1Dpmu_nu";
fPlotTitles = "; p_{#mu} (GeV/c); d#sigma/dp_{#mu} (cm^{2}/(GeV/c)/nucleon)";
EnuMin = 0.;
EnuMax = 100.;
Measurement1D::SetupMeasurement(inputfile, type, rw, fakeDataFile);
// Data comes in ROOT file
// hResultTot is cross-section with all errors
// hResultStat is cross-section with stats-only errors
// hTruthNEUT is the NEUT cross-section given by experimenter
// hTruthGENIE is the GENIE cross-section given by experimenter
SetDataFromFile(GeneralUtils::GetTopLevelDir()+
"/data/T2K/CC1pip/H2O/nd280data-numu-cc1pi-xs-on-h2o-2015.root",
"MuMom/hResultTot");
SetCovarFromDataFile(GeneralUtils::GetTopLevelDir()+
"/data/T2K/CC1pip/H2O/nd280data-numu-cc1pi-xs-on-h2o-2015.root",
"MuMom/TotalCovariance", true);
SetupDefaultHist();
fScaleFactor = (fEventHist->Integral("width")*1E-38)
/double(fNEvents)/TotalIntegratedFlux("width");
};
Specifying the event-level dependent variable(s)
We are now at the point where we can write the FillEventVariables(FitEvent *event) implementation mentioned above. This function defines the dependent variable(s) and how to get them from each FitEvent object that gets passed.
The FitEvent class provides numerous getter functions to make FillEventVariables implementation as simple as possible.
This is the only major point where the pμ and Erecν implementations differ.
For the pµ distribution
To get the muon momentum, we first need a muon in the event. The function FitEvent::NumFSParticle(int pdg) counts the number of final-state particles with pdg-code pdg and returns that number. So our first check is
// Need to make sure there's a muon if (event->NumFSParticle(13) == 0) return;
To then get the muon kinematics in the FitEvent we use FitEvent::GetHMFSParticle(int pdg) to get the HighestMomentumFinalState particle, which return a FitParticle object. The implementation for FitParticle is in FitBase/FitParticle.cxx. We can now access the ROOT object TLorentzVector in the FitParticle which stores the kinematics for the muon. So after we know there's a muon in the event we get the muon by:
// Get the muon TLorentzVector Pmu = event->GetHMFSParticle(13)->fP;
Now we just need the momentum of this TLorentzVector, which can simply be done by calling TLorentzVector::Vect()::Mag(), amongst others.
However, we recommend using the FitUtils namespace to unify the implementations and minimize errors. The FitUtils namespace lives in src/Utils/FitUtils.cxx, in which we see the double p(TLorentzVector) function.
To get the muon momentum (in GeV) we do
double p_mu = FitUtils::p(Pmu);
and then to finally set the Measurement1D member which saves the dependent variable we do
fXVar = p_mu; return;
which concludes the FillEventVariables(FitEvent *event) function.
For the Erecν distribution
Erecν needs to be reconstructed from the outgoing particles, so requires a little more work. However it follows very similar steps to the muon case.
To reconstruct Erecν we need:
- Muon momentum
- Pion momentum
- Muon/neutrino angle
- Pion/neutrino angle
- Muon/pion angle
- All the particle masses
So we proceed to find the TLorentzVectors in the event as before. We first need to make sure we have a positive pion and a negative muon in the event:
// Need to make sure there's a muon if (event->NumFSParticle(13) == 0) return; // Need to make sure there's a pion if (event->NumFSParticle(211) == 0) return;
Then get the relevant TLorentzVectors:
// Get the incoming neutrino TLorentzVector Pnu = event->GetNeutrinoIn()->fP; // Get the muon TLorentzVector Pmu = event->GetHMFSParticle(13)->fP; // Get the pion TLorentzVector Ppip = event->GetHMFSParticle(211)->fP;
We should now write a helper function in the FitUtils namespace to get Erecν from the muon, pion and neutrino TLorentzVectors. The paper specifies the exact form in equation 1. The FitUtils implementation lives in src/Utils/FitUtils.cxx.
double FitUtils::EnuCC1piprec(TLorentzVector pnu, TLorentzVector pmu, TLorentzVector ppi) {
// The muon energy, momentum and mass
double E_mu = pmu.E() / 1000.;
double p_mu = pmu.Vect().Mag() / 1000.;
double m_mu = sqrt(E_mu * E_mu - p_mu * p_mu);
// The pion energy, momentum and mass
double E_pi = ppi.E() / 1000.;
double p_pi = ppi.Vect().Mag() / 1000.;
double m_pi = sqrt(E_pi * E_pi - p_pi * p_pi);
// The relevant angles
double th_nu_pi = pnu.Vect().Angle(ppi.Vect());
double th_nu_mu = pnu.Vect().Angle(pmu.Vect());
double th_pi_mu = ppi.Vect().Angle(pmu.Vect());
// Now write down the derived neutrino energy
// assuming the initial state nucleon is at rest
// and we only have a pion, muon and nucleon in the final state
double rEnu = (m_mu * m_mu + m_pi * m_pi - 2 * m_n * (E_pi + E_mu) + 2 * E_pi * E_mu -
2 * p_pi * p_mu * cos(th_pi_mu)) /
(2 * (E_pi + E_mu - p_pi * cos(th_nu_pi) - p_mu * cos(th_nu_mu) - m_n));
return rEnu;
};
Once we have the helper function implemented and tested we go back to the FillEventVariables(FitEvent *event) implementation and simply do
double Enu = FitUtils::EnuCC1piprec(Pnu, Pmu, Ppip); fXVar = Enu; return;
and FillEventVariables(FitEvent *event) is done!
Specifying a signal definition
The implementation of our new sample now just needs a signal definition.
The base class function MeasurementBase::Reconfigure() specifies isSignal(FitEvent *) which is what we will implement in our class. Since we plan on re-using the signal definition for multiple distributions, we should include it in the src/T2K/T2K_SignalDef.cxx implementation, as mentioned earlier.
Reading the paper, we see the two requirements:
"The CC1π+cross section is described by the particles leaving the nucleus, i.e. one muon, one positive pion and any number of nucleons" (
and
"The analysis presented restricts the kinematic phase-space to the region defined by pµ > 200 MeV/c, pπ+ > 200 MeV/c, cos(θµ) > 0.3 and cos(θπ+ ) > 0.3."
so our signal definition needs to extract these quantities from the FitEvent object and perform the selection upon them.
We prefer a clear unambiguous naming scheme, so let's name the namespace function isCC1pip_T2K_H2O. The function will need to take the FitEvent object along with the EnuMin and EnuMax defined in the experiment signal definition.
We can use already declared functions of the namespace SignalDef: these live in src/Utils/SignalDef.cxx. We can see a SignalDef::isCC1pi(FitEvent *, int nuPDG, int piPDG, double EnuMin, double EnuMax), which looks promising. This helper function searches for:
- 1 single lepton of type
nuPDG-1 - 1 single pion of type
piPDG - makes sure the neutrino energy is between
EnuMinandEnuMax
We can then add our kinematic phase-space requirements on the muon and pion in our SignalDef::isCC1pip_T2K_H2O(FitEvent *event, double EnuMin, double EnuMax implementation in src/T2K/T2K_SignalDef.cxx.
What to do:
We get for src/T2K/T2K_SignalDef.cxx:
namespace SignalDef {
// T2K H2O signal definition
bool isCC1pip_T2K_H2O(FitEvent *event, double EnuMin,
double EnuMax) {
if (!isCC1pi(event, 14, 211, EnuMin, EnuMax)) return false;
TLorentzVector Pnu = event->GetHMISParticle(14)->fP; // Can also use FitEvent::GetNeutrinoIn()
TLorentzVector Pmu = event->GetHMFSParticle(13)->fP;
TLorentzVector Ppip = event->GetHMFSParticle(211)->fP;
double p_mu = FitUtils::p(Pmu) * 1000;
double p_pi = FitUtils::p(Ppip) * 1000;
double cos_th_mu = cos(FitUtils::th(Pnu, Pmu));
double cos_th_pi = cos(FitUtils::th(Pnu, Ppip));
if (p_mu <= 200 || p_pi <= 200 || cos_th_mu <= 0.3 || cos_th_pi <= 0.3) {
return false;
}
return true;
};
}
Then back in src/T2K/T2K_CC1pip_H2O_XSec_1Dpmu_nu.cxx we simply do:
//********************************************************************
// Beware: The H2O analysis has different signal definition to the CH analysis!
bool T2K_CC1pip_H2O_XSec_1Dpmu_nu::isSignal(FitEvent *event) {
//********************************************************************
return SignalDef::isCC1pip_T2K_H2O(event, EnuMin, EnuMax);
}
making sure our header file has #include "T2K_SignalDef.h".
Add the sample to the CMake process
CMake is incredibly easy and versatile and the only modification you need to make is in src/T2K/CMakeLists.txt.
You simply need to add the implementation files to the set(IMPLFILES list and the header files to the set(HEADERFILES.
What to do:
- Add
T2K_CC1pip_H2O_XSec_1Dpmu_nu.cxxandT2K_CC1pip_H2O_XSec_1DEnuMB_nu.cxxtoset(IMPLFILES
- Add
T2K_CC1pip_H2O_XSec_1Dpmu_nu.handT2K_CC1pip_H2O_XSec_1DEnuMB_nu.htoset(HEADERFILES
Make NUISANCE aware of the sample
Now we've got all we need to include the sample in NUISANCE. The full implementation is listed at the bottom of this page for reference.
As mentioned earlier, NUISANCE loads samples through src/FCN/SampleList.cxx and specifically bool SampleUtils::LoadSample. We need to add our new sample to this long if else statement, making sure we string compare to the name set previously.
What to do:
Add in
/*
T2K CC1pi+ H2O samples
*/
} else if (!name.compare("T2K_CC1pip_H2O_XSec_1DEnuMB_nu")) {
fChain->push_back(new T2K_CC1pip_H2O_XSec_1DEnuMB_nu(file, rw, type, fkdt));
} else if (!name.compare("T2K_CC1pip_H2O_XSec_1Dpmu_nu")) {
fChain->push_back(new T2K_CC1pip_H2O_XSec_1Dpmu_nu(file, rw, type, fkdt));
}
after the last T2K entry.
Also add the relevant header files for the new samples in the src/FCN/SampleList.h:
// T2K CC1pi+ on H2O #include "T2K_CC1pip_H2O_XSec_1DEnuMB_nu.h" #include "T2K_CC1pip_H2O_XSec_1Dpmu_nu.h"
Recompiling NUISANCE with the new sample
Since we've added files to the CMakeList.txt we need to reprocess the makefiles produced by CMake.
What to do:
$ mkdir -p build $ cd build $ cmake ..
We can then proceed as usual by cleaning the make, sourcing our environment and making the project:
$ make clean $ source $uname/setup.sh $ make -j $ make docs $ make install
If all is well we exit without any errors and we can proceed to making predictions for our new sample in NUISANCE.
Running NUISANCE with the new sample
We can now write a card file and run any of the NUISANCE application with the sample.
If you haven't got any generated MC, now's the time to generate them! Alternatively, check our generated MC for T2K ND280 flux with H2O target; it should be available for most generators.
What to do: Once you have your sample make a NUISANCE input card file, e.g.
$ vim my_T2KH2O_card.card
and specify the sample in the card file as
sample T2K_CC1pip_H2O_XSec_1Dpmu_nu GENERATOR:PATH_TO_GENERATOR_OUTPUT_FILE
where you replace GENERATOR with whatever generator you ran with, all in caps (e.g. GENIE, NEUT, NUWRO, GIBUU or NUANCE).
Then finally run any of the NUISANCE executables, specifying the input card file and output root file e.g.
$ ./nuiscomp -c my_T2KH2O_card.card -o testing_my_T2KH2O_impl.root
Hot tip: If you're testing/debugging an implementation you might not want to run over several million events. You can change the number of loaded events by appending -q input.maxevents=50000 to run with 50000 events instead.
The final implementation
Here's a summary of the implementation for reference:
T2K_CC1pip_H2O_XSec_1Dpmu_nu.cxx:
#include "T2K_CC1pip_H2O_XSec_1Dpmu_nu.h" // The muon momentum //******************************************************************** T2K_CC1pip_H2O_XSec_1Dpmu_nu::T2K_CC1pip_H2O_XSec_1Dpmu_nu( std::string inputfile, FitWeight *rw, std::string type, std::string fakeDataFile){ //******************************************************************** fName = "T2K_CC1pip_H2O_XSec_1Dpmu_nu"; fPlotTitles = "; p_{#mu} (GeV/c); d#sigma/dp_{#mu} (cm^{2}/(GeV/c)/nucleon)"; EnuMin = 0.; EnuMax = 10.; Measurement1D::SetupMeasurement(inputfile, type, rw, fakeDataFile); // Data comes in ROOT file // hResultTot is cross-section with all errors // hResultStat is cross-section with stats-only errors // hTruthNEUT is the NEUT cross-section given by experimenter // hTruthGENIE is the GENIE cross-section given by experimenter SetDataFromFile(GeneralUtils::GetTopLevelDir()+ "/data/T2K/CC1pip/H2O/nd280data-numu-cc1pi-xs-on-h2o-2015.root", "MuMom/hResultTot"); SetCovarFromDataFile(GeneralUtils::GetTopLevelDir()+ "/data/T2K/CC1pip/H2O/nd280data-numu-cc1pi-xs-on-h2o-2015.root", "MuMom/TotalCovariance", true); SetupDefaultHist(); fScaleFactor = (fEventHist->Integral("width")*1E-38)/double(fNEvents)/TotalIntegratedFlux("width"); }; //******************************************************************** // Find the momentum of the muon void T2K_CC1pip_H2O_XSec_1Dpmu_nu::FillEventVariables(FitEvent *event) { //******************************************************************** // Need to make sure there's a muon if (event->NumFSParticle(13) == 0) return; // Get the muon TLorentzVector Pmu = event->GetHMFSParticle(13)->fP; double p_mu = FitUtils::p(Pmu); fXVar = p_mu; return; }; //******************************************************************** // Beware: The H2O analysis has different signal definition to the CH analysis! bool T2K_CC1pip_H2O_XSec_1Dpmu_nu::isSignal(FitEvent *event) { //******************************************************************** return SignalDef::isCC1pip_T2K_H2O(event, EnuMin, EnuMax); }
T2K_CC1pip_H2O_XSec_1Dpmu_nu.h:
#ifndef T2K_CC1PIP_H2O_XSEC_1DPMU_NU_H_SEEN
#define T2K_CC1PIP_H2O_XSEC_1DPMU_NU_H_SEEN
#include "Measurement1D.h"
#include "T2K_SignalDef.h"
class T2K_CC1pip_H2O_XSec_1Dpmu_nu : public Measurement1D {
public:
T2K_CC1pip_H2O_XSec_1Dpmu_nu(std::string inputfile, FitWeight *rw,
std::string type, std::string fakeDataFile);
virtual ~T2K_CC1pip_H2O_XSec_1Dpmu_nu() {};
void FillEventVariables(FitEvent *event);
bool isSignal(FitEvent *event);
private:
};
#endif
What about 2D distributions?
2D distributions follow a similar implementation pattern: find a suitable set of base-class functions to use in the constructor, specify the FillEventVariables and isSignal, and make additions to the signal definition (SignalDef) and utility (FitUtils) namespaces.
You'll have to add the new sample to the CMake process just like in the 1D case, and add the sample into the SampleList.
Attachments (2)
- T2K_CC1pip_H2O_enurec.png (46.8 KB) - added by 8 years ago.
- T2K_CC1pip_H2O_pmu.png (83.2 KB) - added by 8 years ago.
Download all attachments as: .zip