| Version 14 (modified by , 8 years ago) (diff) |
|---|
How to add a sample in NUISANCE
This is an informal step-by-step guide on how to add samples into the NUISANCE framework.
For this tutorial I'll be adding the T2K CC1pi+ H2O data. The data comes from the T2K site and arxiv. The data is supplied both in a ROOT file and a number of .csv files: I'll be using the ROOT file here.
Versions: NUISANCE v1r0, NEUT 5.3.6, arxiv 1605.07964v2.pdf
Examining the data and choosing distributions
Finding the neutrino flux and generating events
The first issue at hand is to find the flux for the experiment. If we don't have this, we cant make a generator prediction---unless the measurement is a total cross-section without any phase space cuts (in which case you should probably cast a suspicious eye).
A quick search through the arxiv document points us to reference [12]. It is also in our flux list.
We then generate events in NEUT 5.3.6 with a suitable card-file, see our NEUT guide on how to do this. We need the correct target (water) and flux, and perform the model selections we want (e.g. Rein-Sehgal or Berger-Sehgal coherent model). The procedure is very similar for other generators too.
Aim to generate around 1M events with all interaction modes turned on. This way we make sure to get all interaction modes into the topologically defined cross-section. We get a small amount of CCQE events which excite a pion from the outgoing nucleon interacting in the nucleus to kick out a pion, leading to a CCQE+1pi ~ CC1pi+ final state, which is signal for this particular sample.
Choosing a distribution
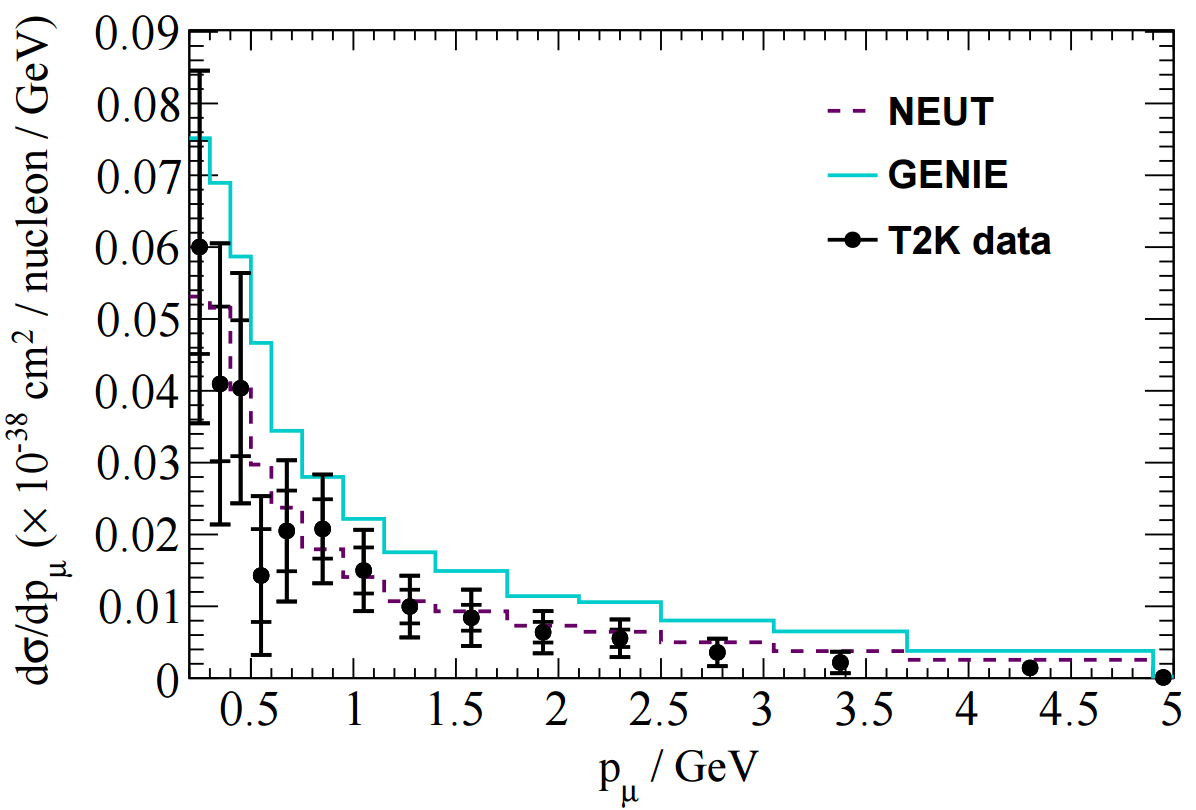
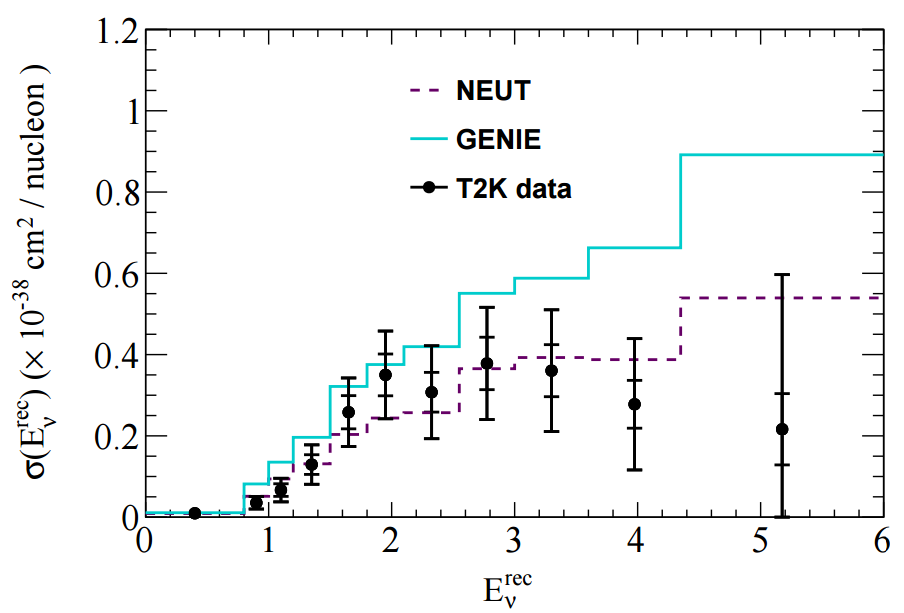
The T2K CC1pi+ H2O data release contains various distributions in FIG 4. In this tutorial I'll look at adding one kinematic distribution and one derived distribution: pmu and Erecnu shown below.
In NUISANCE we try to add all available distributions from a publication. However, some distributions will have detector effects "folded" into them (i.e. they will be raw detector-level data). We can only use these if there is some method which bring detector-level variables (e.g. p_mu seen in the detector) to truth-level variable (e.g. p_mu seen after correcting for the detector effects).
Implementing a sample
NUISANCE is designed to easily allow adding new samples.
Each experimental measurement is its own class. To simplify including new samples we supply a base class for 1D (Measurement1D) and 2D (Measurement2D) measurements, which in turn inherits from the virtual base class (MeasurementBase).
These MeasurementBase classes are then looped over in the executables and data/MC comparisons are the result.
The inheritance tree is simple and goes Specific_Measurement -> MeasurementXD -> MeasurementBase
Setting up the inheritance
In the case of our T2K CC1pi+ H2O data, we're dealing with 1D distributions. They should therefore inherit from the Measurement1D class.
#ifndef T2K_CC1PIP_H2O_XSEC_1DPMU_NU_H_SEEN
#define T2K_CC1PIP_H2O_XSEC_1DPMU_NU_H_SEEN
#include "Measurement1D.h"
#include "T2K_SignalDef.h"
class T2K_CC1pip_H2O_XSec_1Dpmu_nu : public Measurement1D {
public:
T2K_CC1pip_H2O_XSec_1Dpmu_nu(std::string inputfile, FitWeight *rw, std::string type, std::string fakeDataFile);
virtual ~T2K_CC1pip_H2O_XSec_1Dpmu_nu() {};
void FillEventVariables(FitEvent *event);
bool isSignal(FitEvent *event);
private:
};
#endif
Naming the sample
Some automatic processing is done on loading up the sample. Most of these are simple string comparisons, but do place responsibility on the user.
For cross-sections, for event-rates
Placing the sample
Making the constructor
Specifying the event-level dependent variable(s)
Specifying a signal definition
#include "T2K_CC1pip_H2O_XSec_1Dpmu_nu.h" // The muon momentum //******************************************************************** T2K_CC1pip_H2O_XSec_1Dpmu_nu::T2K_CC1pip_H2O_XSec_1Dpmu_nu(std::string inputfile, FitWeight *rw, std::string type, std::string fakeDataFile){ //******************************************************************** fName = "T2K_CC1pip_H2O_XSec_1Dpmu_nu"; fPlotTitles = "; p_{#mu} (GeV/c); d#sigma/dp_{#mu} (cm^{2}/(GeV/c)/nucleon)"; EnuMin = 0.; EnuMax = 10.; Measurement1D::SetupMeasurement(inputfile, type, rw, fakeDataFile); // Data comes in ROOT file // hResultTot is cross-section with all errors // hResultStat is cross-section with stats-only errors // hTruthNEUT is the NEUT cross-section given by experimenter // hTruthGENIE is the GENIE cross-section given by experimenter SetDataFromFile(GeneralUtils::GetTopLevelDir()+"/data/T2K/CC1pip/H2O/nd280data-numu-cc1pi-xs-on-h2o-2015.root","MuMom/hResultTot"); SetCovarFromDataFile(GeneralUtils::GetTopLevelDir()+"/data/T2K/CC1pip/H2O/nd280data-numu-cc1pi-xs-on-h2o-2015.root", "MuMom/TotalCovariance", true); SetupDefaultHist(); fScaleFactor = (fEventHist->Integral("width")*1E-38)/double(fNEvents)/TotalIntegratedFlux("width"); }; //******************************************************************** // Find the momentum of the muon void T2K_CC1pip_H2O_XSec_1Dpmu_nu::FillEventVariables(FitEvent *event) { //******************************************************************** // Need to make sure there's a muon if (event->NumFSParticle(13) == 0) return; // Get the muon TLorentzVector Pmu = event->GetHMFSParticle(13)->fP; double p_mu = FitUtils::p(Pmu); fXVar = p_mu; return; }; //******************************************************************** // Beware: The H2O analysis has different signal definition to the CH analysis! bool T2K_CC1pip_H2O_XSec_1Dpmu_nu::isSignal(FitEvent *event) { //******************************************************************** return SignalDef::isCC1pip_T2K_H2O(event, EnuMin, EnuMax); }
Make NUISANCE aware of the sample
FCN/SampleList
Add the sample to the makefile
CMake
Attachments (2)
- T2K_CC1pip_H2O_enurec.png (46.8 KB) - added by 8 years ago.
- T2K_CC1pip_H2O_pmu.png (83.2 KB) - added by 8 years ago.
Download all attachments as: .zip